Batch pipeline
Purpose

|
The Batch The Batch Pipeline replaces the Batch Filter tool present in previous versions of NeuroInfo. |
Procedure Overview
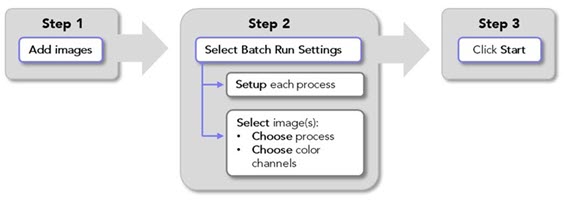
Adding images to the batch pipeline
-
 In the main window, go to the Pipelines ribbon and click Batch Pipeline.
In the main window, go to the Pipelines ribbon and click Batch Pipeline.The Batch Pipeline window opens.
-
Click Add images; the Image Open dialog opens.
Select the images that you want to process; hold the Shift and Ctrl keys on the keyboard to select multiple images.
-
Click Open to add the selected images to the Batch Pipeline.
The images you opened and their file paths will be listed in the Input Image column of the Batch Pipeline window.
Note that, depending on your computer hardware, image stacks with thousands of image planes may not be compatible with processing using the batch pipeline, causing NeuroInfo to hang or freeze.
Buttons
 Load images into the Batch Pipeline
Load images into the Batch Pipeline
 Remove all images from the Batch Pipeline
Remove all images from the Batch Pipeline
 Enable selection of multiple images so that you can set their batch processing settings
Enable selection of multiple images so that you can set their batch processing settings
 Click to start the batch process
Click to start the batch process
Table columns
Note: You can resize the table columns by dragging the vertical dividers
-
 Copy: Create a copy of the image. This is convenient for running different batch processes on a single image, e.g., to evaluate settings in preparation for a large batch processing run.
Copy: Create a copy of the image. This is convenient for running different batch processes on a single image, e.g., to evaluate settings in preparation for a large batch processing run. -
 Delete: Delete the image from the Batch Pipeline.
Delete: Delete the image from the Batch Pipeline. -
Input Image: File names and locations of images that have been added to the Batch Pipeline are displayed
-
Status: Displays "Setup Required" if you need to specify processing settings.
Displays “Ready to start” once the process settings have been selected.
Once the batch process is launched, general information about status is displayed.
-
Detail: Details of the batch-process status are displayed during processing.
-
 Open: Click to open the processed file in the main window.
Open: Click to open the processed file in the main window.
Image Filtering
See also Image filters
(Recommended) Identify appropriate image-filter settings for your images
Before processing a large group of images using the batch pipeline, we recommend that you identify appropriate image-filter settings first. To do this, follow the Image Filtering below to apply the image filter of your choice to one or a few representative images. Here are some tips for doing that:
-
Once you have added your representative image(s) to the batch Pipeline, click
 Copy to create a few copies.
Copy to create a few copies. -
Set up several configurations of the filter that you intend to use with different settings that you want to test.
-
Review the results and identify the settings that provided the best results.
-
Repeat and fine-tune as needed.
Procedure for batch image-filtering
-
Load images for batch processing as described above.
-
Set up/modify image filters if you have not already done so (or go to Step 3 if your image-filters are ready):
-
 In Batch Settings on the right, click the Image Filter Setup button.
In Batch Settings on the right, click the Image Filter Setup button. The Image Filter Setup window opens.
-
 Click Add Configuration to set up a new image filter.
Click Add Configuration to set up a new image filter. To modify an existing image filter, select the saved configuration that you want to change and click Edit Configuration. Note: this will overwrite the existing configuration.
To modify an existing image filter, select the saved configuration that you want to change and click Edit Configuration. Note: this will overwrite the existing configuration. -
Specify a name and settings:
-
Name: Type a name for the filter settings, for example "Golgi default" or "Golgi 10px edge".
-
Filter Type: Select the image filter you want to configure from the drop-down menu.
-
Output Label: A default file-name suffix populates the field; you can change this by typing into the box.
-
Compression: Specify the amount of file compression (if any) to apply to the image using the slider.
-
Settings: Adjust the filter settings. The options depend on the Filter Type selected. See Image filters and settings below for specific information on settings for the available image filters.
-
-
 Click Save Changes to add the new/modified configuration to the Saved Configurations list and make it available for use in the Batch Pipeline.
Click Save Changes to add the new/modified configuration to the Saved Configurations list and make it available for use in the Batch Pipeline. -
 Click Return to list to close the Image Filter Setup window and return to the Batch Pipeline window.
Click Return to list to close the Image Filter Setup window and return to the Batch Pipeline window.
-
-
Select images and choose image-filters and color channels for batch processing:
One image at a time
-
Select an image by clicking its row in the Batch Runs table; the row will be highlighted in blue to indicate that the image is selected.
-
In the Batch Settings section on the right, select an image-filter configuration from the Image filter drop-down menu.
-
Check the boxes to choose the color channel(s) to include in the image-filtering process. If changes have been made to the number of channels being used for detection, click Refresh Display to update the Number of Channels.
Multiple images
-
 Click Select Multiple to enable selection of more than one image.
Click Select Multiple to enable selection of more than one image. -
Select images by clicking their rows or click Select All; blue highlight indicates the images that are selected.
-
In the Batch Settings section on the right, select an image-filter configuration from the Image filter drop-down menu.
-
Check the boxes to choose the color channel(s) to include in the image-filtering process. If changes have been made to the number of channels being used for detection, click Refresh Display to update the Number of Channels.
-
 Click Apply to finish choosing batch pipeline processing settings for the selected files.
Click Apply to finish choosing batch pipeline processing settings for the selected files.
-
-
The Status column for each image loaded into the pipeline should read "Ready to start."
You can view the settings chosen for each image by clicking on it in the Batch Runs table (blue highlight indicates the file that is selected). The chosen processing settings are displayed in Batch Settings on the right.
-
 Click Start to launch the batch process.
Click Start to launch the batch process.-
The Detail column displays the current status of the process.
-
 When processing is complete, the Status column will read "Completed" and you can view the results by clicking the open-file icon in the Open column.
When processing is complete, the Status column will read "Completed" and you can view the results by clicking the open-file icon in the Open column.
-
Image filters and settings
Golgi: Reduce out-of-focus signal in brightfield microscopy.
The Golgi filter cuts down the out-of-focus shadows that are the result of labeled structures in the tissue. It works for solidly labeled objects such as those produced by Golgi staining and cannot be used for improving the appearance of very transparent or weakly-labeled structures, nor for improving the appearance of small blurry objects.
Settings
Compression: Use the slider to indicate your desired file-compression ratio.
Sensitivity: Used to control the amount of signal necessary for an object to be selected as part of the foreground structure. To capture weakly-labeled structures, you can use values greater than 100%.
Edge clearance: Used to mitigate edge artifact that may occur if the algorithm takes into account information from the edge of the image.
With edge clearance >0, the algorithm processes the edge, but without using contrast information within the specified number of pixels around the image.
Brightfield image: Leave the box checked if you're working with a brightfield microscopy image.
In vivo: Reduce motion artifacts in images from live specimens.
Consecutive in vivo-acquired images in a stack may appear slightly shifted and distorted at high resolution due to blood flow in and around the field of view. The in vivo filter is designed to reduce/remove these motion artifacts.
Settings
Maximum displacement: Represents the maximum number of pixels that a particular feature can move from frame to frame in any direction.
Feature radius (px): Represents the radius size (in pixels) of the image landmarks used for estimating displacement.
Minimum feature value: Value used for landmark detection. The higher the value, the fewer landmarks detected.
Closing: Perform a grayscale morphological closing of the image; useful for closing small gaps inside a labeled structure.
Settings
Structuring radius: Radius size of the gap that the filter is able to close.
Brightfield Image: Check the box to configure the filter for brightfield images. Leave it unchecked to configure for fluorescence images.
Projection: Maximum and Minimum projection display an entire image stack in a single plane. To display the projection, NeuroInfo software identifies the maximum (or minimum) intensity value of each pixel along the Z axis for projection onto the 2D plane.
Settings
Intensity Projection Settings:
-
Select Max intensity projection with light-on-dark images (e.g., fluorescence)
-
Select Min intensity projection with dark-on-light images (e.g., brightfield).
Vessel: Make vessels and other hollow structures appear solid.
The Vessel filter identifies the boundaries of labeled structures and then synthesizes a cross-section of even intensity based on a model adjusted to fit the input data. An evenly-labeled foreground is generated in the output image, which is more suitable for digital reconstruction.
Settings
Sensitivity: Control the amount of signal necessary for an object to be selected as part of the foreground structure. To capture weakly-labeled structures, you can use values greater than 100%.
Edge clearance: Helps ensure that 'holes' in the labeled structure that are adjacent to the edge of the image are properly filled. Holes that extend into the image beyond the edge clearance value (in pixels) are treated as background and are not filled in.
Sharpen Image: Reduce edge blurriness by enhancing edges using the unsharp mask technique.
Notes: Sharpening enhances edges in images using a local adjustment of image contrast. Sharpening can enhance image noise, we recommend applying minimal image sharpening to avoid this.
Changes in image appearance after sharpening are subtle; you may have to zoom in to see the changes.
With RGB images, you must ungroup color channels in the Image Adjustment Panel before applying the filter.
Settings
-
Scale (µm): The size of the edges to sharpen. For most images, we suggest starting with 1–2 µm.
-
Amount (%): The magnitude of the image-contrast adjustment.
Scale and Amount settings affect each other; reducing one generally makes it possible to increase the other.
-
Threshold levels: The minimum difference between adjacent brightness levels needed for the filter to make an adjustment. The threshold setting can help you avoid sharpening weak edges and making smooth areas appear speckled.
Background subtraction: Minimize background in images by estimating and removing background illumination using an adaptive, local adjustment of image contrast.
Note that, in addition to removing background in individual images, this filter can also reduce tiling artifacts seen in large image montages if it is applied to the individual tiles before assembly.
Settings
Diameter (µm): The physical scale of the foreground objects to preserve. In most cases, this value should be larger than the size of the objects you want to visualize. If this parameter is set too low (small), the centers of larger objects may be removed. If it is set too high, the filtering may not have the desired impact on image quality.
ExM: Make labeled structures in images of expanded tissue appear solid.
The ExM filter changes the appearance of labeled structures in images of expanded tissue from their typical punctate appearance into solidly labeled structures. The filter uses a pre-trained Machine Learning (ML) model to replace the selected channel with a foreground probability map based on the density of puncta in specific regions. The resulting foreground probability map can then be used to perform morphologic reconstructions using available tracing and detection algorithms. To obtain the best results the image black point and white point should be properly adjusted and saved to file prior to processing.
Settings
Compression: Use the slider to indicate your desired file-compression ratio.
Sample radius: Controls the size of the 3D image patches that are processed. The ExM filter uses 3D image patches that are 128 pixels across. Therefore, to use 1:1 sampling you simply multiply the lateral resolution of your images by 64. Larger values result in coarser predictions.
Segm. Model: Select one of the pre-trained ML models to compute the foreground probability.
Tone mapping: Apply localized contrast adjustments to reveal details in both very bright and very dark regions of an image.
Tone mapping applies local contrast adjustments that make it easier to visualize details in both bright and dark regions of an image simultaneously. It can be useful for exploring large images that exhibit a very high dynamic range between the brightest and darkest parts of the image.
The filter is appropriate for use with bit-depth images (10-bit or greater).
Tone mapping can be applied to one or more channels of the image. One common use is to better visualize neurons in tract-tracing experiments where the injection site is very bright compared to the regions of the brain to which the neurons project. In this case, tone mapping could be applied to the color channels used for tract tracing, but not applied to the color channel that labels cytoarchitecture.
How it works: Tone mapping separates images into a base layer that encodes the large-scale intensity variations and a detail layer that encodes small-scale details. Contrast adjustments are made to the base layer to smooth the large intensity changes, and the result is recombined with the detail layer.
Settings