Using Optical Fractionator with external image sources
Purpose
Use this modified Optical Fractionator workflow to count objects in image stacks acquired in a systematic random sampling (SRS) manner using another microscope system (i.e., not an MBF Bioscience image acquisition system).
To ensure an unbiased estimate, images must be acquired in a systematic way with equal spacing in the X and Y axes between all the images throughout the entire region of interest on a section. The same spacing needs to be used to acquire images on all sections that contain the region of interest in order to get an estimate for the entire structure. The distance between images will be used when setting up the grid layout in the workflow.
If you are counting after acquiring image stacks using the SRS image stack series workflow in Stereo Investigator, see Using Optical Fractionator to count offline after an SRS Image Stack acquisition .
Procedure
-
 Click the Optical Fractionator workflow button on the Probes ribbon.
Click the Optical Fractionator workflow button on the Probes ribbon. If you haven't used the workflow recently, find the Optical Fractionator workflow button in the Number drop-down menu in the All Probes section of the Probes ribbon.
If you haven't used the workflow recently, find the Optical Fractionator workflow button in the Number drop-down menu in the All Probes section of the Probes ribbon. -
In the dialog box that opens, choose Start a new subject from image stacks.
-
Click OK to close the Optical Fractionator Workflow dialog box. The workflow panel is displayed.
-
Follow the steps in the workflow.
Using the workflow
-
Subject Information: Type your name, the subject of the study, and any notes pertaining to this study or animal.
-
Use Saved Sampling Parameters: Choose yes if you want to use a previously saved sampling configuration.
-
If yes, click the browse
 icon.
icon. -
In the Sampling Parameter Chooser window that opens, highlight the parameter set you wish to use and click OK.
-
-
Enter Serial Section information:
-
# of sections to count: Number of sections you will count for this subject across all of the slides.
-
Section cut thickness (Required to move to the next step): Tissue cut thickness or block advance.
-
Section evaluation interval: Interval at which you want to count serially cut sections. For example, if every third tissue section will be counted, enter 3.
-
Starting section number: Number of the first section to be counted.
-
Randomize button: If you are counting in multiple sections, we recommend clicking the Randomize button so that the first sampling section is randomly selected.
-
If you don't want to keep track of the physical section count, leave this number as "1."
-
-
Z-value of first section: This number is automatically adjusted based on the Starting Section Number entered; it reflects an interval of the section cut thickness.
-
To modify the thickness, click the Edit Serial Sections button.
-
-
Load images by section: From the dropdown menu, select the section you want to load images for.
-
Select folder containing images: Click Browse to navigate the folder containing the images. Highlight the folder and click Select Folder.The images are listed below the dropdown menu.
-
Enter grid size used to capture images:
-
Enter the distance in X and Y between the images when they were acquired.
-
Choose a contour from the dropdown menu. We recommend that the contour is named after the region of interest to simplify viewing results. To change contour names, click the cog icon
 in the Contours section of the Trace ribbon in the main Stereo Investigator window.
in the Contours section of the Trace ribbon in the main Stereo Investigator window. -
Click Setup Grid. The images will load in grid formation.
-
-
Measure the mounted thickness while counting is recommended at every sampling site (or at least an evaluation interval) to ensure an unbiased estimation.
-
If you select Manually enter the average mounted thickness, enter the Average Mounted Thickness, under Manual Adjustment.
-
If you select Measure mounted thickness before counting, click the Start Taking Measurements button below to measure the section thickness at several sites prior to counting.
If you only collected images for the disector height, enter the section thickness after shrinkage manually. Keep in mind that, if you enter an incorrect thickness value, you are introducing sampling error and bias to the estimation.
-
Counting Frame Display:
-
Force the counting frame to be square: Check the box to create a square counting frame as you adjust the frame with your mouse.
-
Snap to increments of: Check the box and enter the desired rounding increment in µm to round the size of the counting frame to a whole number; this may be easier to remember for future experiments.
-
-
Counting Frame Size: Size the counting frame to fit approximately 1–5 objects of interest.
-
Choose a unique identifying point for your object of interest (e.g., cell) that comes into focus just once, such as:
-
Cell top
-
Nucleus top
-
Nucleolus (unless there are multiple nucleoli in your cells of interest)
-
-
Adjust the counting frame until it is approximately large enough to have, on average, 1 to 5 identifying points that you will count using one of the following methods:
-
Type a size (µm) into the boxes labeled X: and Y:.
-
Use your mouse to adjust the counting frame size.
We recommend sizing counting frames so that they contain 1–5 objects. This minimizes user error and fatigue; it is difficult to count too many objects per frame, especially in a visually dense image.
-
-
Verify the following:
-
The counting frame is roughly in the center of the screen. Hover your cursor over the counting frame and drag the mouse to move the frame if needed.
-
There is adequate space outside the counting frame to clearly distinguish objects that are on the edge of the counting frame.
-
The tails of the counting frame are visible.
-
-
-
Optional: Add a second counting frame
You can use a second counting frame to mark a population of objects that require different parameters than the objects being counted in the primary counting frame. This allows you mark objects with both sets of parameters in a single probe run which can save time. To use a second counting frame, you CTRL-click left mouse button to count/mark objects. When enabled, you can count a second object in the second frame using middle mouse button CTRL-click.
The second counting frame is useful for counting objects that are different in:
-
Size: You can set the main frame-size to accommodate 1–5 of the larger cells and add a second, smaller counting frame appropriate for 1–5 of the smaller cells.
-
Population density: Use the main counting frame for the more sparsely distributed objects and set the second counting frame with an appropriate interval to count the densely distributed objects.
Add Second Counting Frame: Check the box to add a counting frame inside the primary counting frame. The following options for the second counting frame will become available:
-
Centered check box:
- When checked, the second counting frame is centered in the primary counting frame.
- When unchecked, the second counting frame is positioned at the top left inside the primary counting frame.
-
Interval: Type in the interval at which you want a second counting frame. For example, enter "1" to include a second counting frame in each primary counting frame or enter "5" to have a second counting frame every fifth primary counting frame.
Choose a radio button to indicate how you want to indicate the size of the second counting frame:
-
Set CF dimensions: Select this option and text boxes will appear for you to type in the X and Y dimensions of the second counting frame (in µm).
-
Define as a percentage of primary CF: Select this option and enter a percentage.
To associate a marker with the second counting frame, you must hold the CTRL key down on the keyboard when placing the marker, otherwise the marker will be associated with the first counting frame even if it is contained within the area of the second counting frame.
-
-
Optical disector settings:
-
Check the box to Enter the Guard Zone Heights as Percentages to use percentages of the sample depth for guard zones, rather than using absolute values.
-
Enter the guard zone and disector heights using one of the following options in the dropdown menu:
- Enter top guard zone and disector height; the bottom guard zone is set as the remainder.
- Center the disector between the guard zones. See Guard Zones for more information.
-
-
Focus method:Choose how you want to focus on the sample for counting.
-
Manual Focus: Focus manually: this works best for most people.
-
Automatic Focus (Scanning upwards): The system scrolls the focus from bottom to top automatically.
-
Automatic Focus (Scanning downwards): The system scrolls the focus from top to bottom automatically.
You may want to try using one of the Automatic Focus choices if your counting operations can be completed rapidly. -
-
Sampling Parameter Set: To save the sampling parameters defined in the previous steps:
- Enter a Name for the parameters and any comments you want to include.
- Click Save your Current Settings.
You can use the saved sampling parameters for subsequent specimens by selecting them in Step 1. Set up the subject.
DO NOT change parameters for every section within a specimen. All parameters must be kept constant throughout all the sections of the specimen for the calculations to be valid.
-
Current Sampling Parameter Settings: Displays the sampling parameters that were chosen in previous steps and what will be used for counting.
-
Under Regions of Interest, expand Section 1 by clicking the + sign, expand the region of interest by clicking the + sign, and highlight Acquired Stacks.
-
Click the Play button to start counting. The stack is loaded and you are directed to the first sampling site.
-
The Focus Top of Section dialog box is displayed. Using the PageUp key on the keyboard, focus to the top of the stack, then click OK in the dialog box.
-
If applicable, the Focus Bottom of Section dialog box is displayed. Using the PageDown key, focus to the bottom of the stack, then click OK.
-
Select a marker from the Use Marker drop-down menu in the workflow panel, or from the marker toolbar.
-
Count the cells following the counting rules.
-
Click the fast forward button to go to the next site.
-
Repeat step 7.a–7.g until all sites have been visited.
-
Click Begin Next Section or Add New Section if a new section needs to be added. If you add a new section, the workflow will redirect you step 2. Load images into grid layout.
-
When all sections have been counted, click I’m Finished Counting, or Next Step, to move to Step 8. View sampling results.
Note: It can sometimes be easier to use the mouse wheel to focus through the image stacks rather than use the Page up/down keys on the keyboard. You can setup the mouse wheel focus in the Movement section in Preferences.
Turn on (check) focus with mouse wheel and set the Z distance per wheel click equal to the distance between images in your image stack. 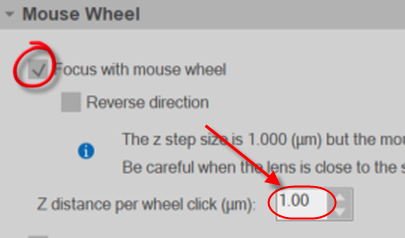
View the results using one of the following methods:
-
Click a set in the Probe Runs list to select it and click the View Results button. The Sampling Results window is displayed.
-
Click Display Probe Run List (use after running more than one probe if you want results from multiple probe runs). The Previous Stereological Runs window is displayed.
-
From the list, highlight the probe runs of interest.
To view results for an entire region of interest, Ctrl-click to select all probe runs from the sections containing the region of interest. This will generate results for the entire structure, not just one section of the structure.
-
Click View Results. The Sampling Results window is displayed.
-
Click on a category in the left hand panel to display the corresponding results in the right hand panel.
-
Optional: Click Export to export all results directly to Excel (2003 or later).
-
-
To see the estimated total cell numbers, highlight the marker name in the left hand panel.
-
Estimated Population using User Defined Section Thickness: Calculated using the thickness manually entered prior to counting the cells.
-
Use this number ONLY if you have measured the thickness separately, before counting.
-
Equals zero if you didn't enter the thickness value manually.
-
-
Estimated Population using Mean Section Thickness: Calculated using the average thickness of the tissue measured at every site, whether there are marked cells or not.
-
Estimated Population using Mean Section Thickness with Counts: Calculated using the average thickness of the tissue measured only at sites containing marked cells.
-
Estimated Population using Number Weighted Section Thickness: Calculated using the thickness of the tissue measured only at sites containing marked cells. These measured thickness values are then weighted by the number of objects associated with them to produce a weighted average. The estimate takes into account wavy tissue with large variation in thickness between sampling sites.
-
Click here for a more detailed description of Optical Fractionator results.