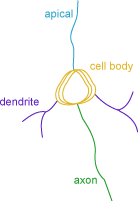
Neuron summary (branched structure analysis)
The Neuron Summary provides an overview of the selected neuron components: axons, dendrites, and cell body.
|
|
The Neuron Summary includes lengths, areas, volumes, quantities, and complexity. Quantities include the number of nodes, terminations, and spines on the branched structures. |
Analysis results
Neuron Summary
Structure names are assigned based on the tool selected for tracing/detection. Some modifications can be specified in Neurolucida or Neurolucida 360 tracing preferences.
For tree structures, the numbers of nodes, ends, and spines are reported. If spines have been classified to identify spine type, numbers of specific spine types are reported.
- Cell body: Total perimeter for all cell bodies included in the analysis.
- Dendrites/Axons: Total length for all branched structures of a given type.
The surface area is modeled as a series of cylindrical sections capped by the end profiles.
Surface Area per Cylinder = [Perimeter of the profile] * [Distance to the next profile]
Calculated for 2+ cell bodies.
|
The calculations are based on this principle: Two stacks with the same number of sections and the same thickness for each section have the same volume. |

|
- Cell body: [Area of the profile] * [Distance to the next profile] — (only available for 2+ cell bodies)
- Dendrite/axon: Computed by modeling each piece of each branch as a frustum.
|
The calculations for the cell body are based on this principle: Two stacks with the same number of sections and the same thickness for each section have the same volume. |

|
In Neurolucida Explorer, complexity refers to the normalization and comparison of dendrites among fundamentally different neurons.
Complexity = [Sum of the terminal orders + Number of terminals] * [Total dendritic length / Number of primary dendrites]
Terminal: Refers to endings
Terminal order: Number of "sister" branches encountered as you proceed from the terminal to the cell body (calculated for each terminal).
Complexity is derived from the Dendritic Complexity Index described in Dendritic Morphology of Hippocampal and Amygdalar Neurons in Adolescent Mice Is Resilient to Genetic Differences in Stress Reactivity (Pillai, de Jong, Kanatsu and al., 2012).
If there is more than one object of a given type, Neurolucida Explorer generates the mean for Length, Area, Surface, Volume.
The area refers to 2-dimensional data while surface refers to 3-dimensional data.
Cell Body Contours
Note that somas detected automatically in the Neurolucida 360 3D environment are contoured in X-Y at each z-plane in which the cell appears. Typically multiple contours will be reported for each soma and measurements for each contour are reported.
Length of the contour representing the cell body.
The 2-dimensional cross-sectional area contained within the boundary of the cell body.
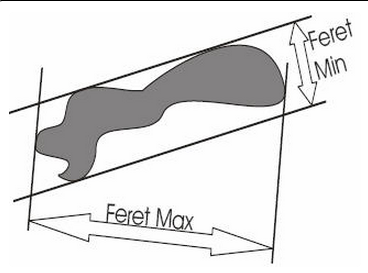
Feret maximum and Feret minimum refer to the largest and smallest dimensions of a contour, respectively, as if a caliper were used for measurement. The two measurements are independent of one another and not necessarily at right angles to each other.
Feret maximum and Feret minimum are reported for each contour drawn.
Aspect Ratio
Aspect ratio = Feret Maximum ÷ Feret Minimum
The minimum aspect ratio is 1. A circle has an aspect ratio of 1, a square approximately 1.4.
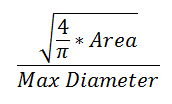
|
|
Convexity = [Convex Perimeter] / [Perimeter]
- A completely convex object does not have indentations, and has a convexity value of 1 (e.g., circles, ellipses, and squares).
- Concave objects have convexity values less than 1.
- Contours with low convexity have a large boundary between inside and outside areas.

|
The form factor differs from the compactness by considering the complexity of the perimeter of the object. For example, a circle with a smooth perimeter has a compactness of 1 and a form factor of 1. If the smooth perimeter is replaced with a finely jagged edge (like a cell covered in microvilli), the compactness is still near 1, but the form factor is much smaller since the perimeter is lengthened considerably.
|
Roundness = [Compactness]2
Use to differentiate objects that have small compactness values.
Solidity = [Area] / [Convex Area]
The area enclosed by a ‘rubber band’ stretched around a contour is called the convex area.
- Circles, squares, and ellipses have a solidity of 1.
- Indentations in the contour take area away from the convex area, decreasing the actual area within the contour.
It is possible to have contours with low convexity and high solidity, and vice versa.
The calculations for cell body are similar to the calculations used for contours: see Contour Measurements in the Neurolucida user guide.
Cell bodies summary
Unless specified in contour settings, soma are assigned a number, e.g., Soma 1, Soma 2.
The number of contours used to define the cell body/bodies. Somas detected automatically in the Neurolucida 360 3D environment are contoured in X-Y at each z-plane in which the cell appears; typically multiple contours will be reported for each soma.
Volume of the cell body contour (2D) or of the shell made from the cell body contours (3D)
Coordinates (x, y, z) of the centroid or geometric center of the cell body