Advanced settings for automatic tree tracing (3D)
See Tracing trees in automatic mode (3D) for step-by-step instructions.
Seed detection tab
Sampling density: The program applies an invisible grid to sample the image uniformly at the density you specify by clicking in the slider:
- Coarse: Fewer grid lines and less sampling performed; this can result in missing vessels.
- Medium: Optimizes density with speed, seed coverage, and final reconstruction accuracy.
- Dense: More grid lines and more sampling are performed. Dense sampling typically leads to longer processing times to produce more seeds and increase the tracing coverage. Accuracy may not improve with this setting; it may result in seeds placed in the background where no vessels exist.
Tracing tab
Quick set
Use the drop-down menu to select from a list of image types that most closely match your image. This will auto-adjust the settings below to our recommended starting point for the advanced settings.
Detector Options
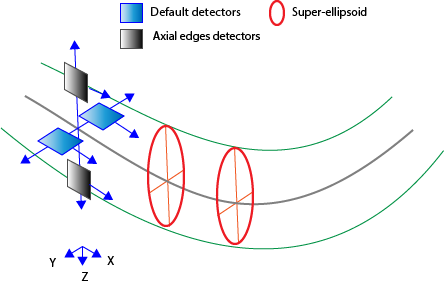
Detect axial edges: Check this box to enable detectors that are Z-aligned. These detectors are in addition to two default detectors that can move left, right, up, and down.
For best results, we recommend enabling this option for most images, but keeping it off for brightfield images.
Use super-ellipsoids with __ iterations: In addition to using detectors, it is possible to fit and model super-ellipsoids at each tracing point.
- This usually gives better XYZ location and diameter estimations, and requires relatively wide tree (in pixels).
- The number of iterations affect the accuracy in fitting super-ellipsoids; more iterations will provide better accuracy; however, the analysis will typically take more time.
-
When Confocal: High Magnification is the selected Quick set, an additional drop-down appears so that you can specify whether the chosen number of iterations are used "As Needed" or "Always". The "As Needed" setting may speed the analysis by directing the fitting to halt when additional iterations would not enhance accuracy.
Detector Size Range in Pixels
The four (top bottom left and right) detectors have a variable size (in pixels). The longer the detector, the better it responds to gaps, although it might miss curved segments.
Drag the sliders or click the arrows to adjust the range to optimize the tradeoff between detection of gaps and curved segments.
Detector movement constraints
The detectors move in 3D, and rotation and shifting values constrain the movement.
Click in the slider to adjust the detector movement constraints.
Rotation: Maximum 3D turning angle when moving along the processes.
Shifting: Movement inward or outward from the centerline when moving along. For example, if Shifting is set to 3 pixels and the right detector detects an edge 5 pixels away from the centerline, search for the next point will be implemented +/- 3 pixels away from the detected edge, i.e., between 2 and 8 pixels away from the centerline. Shifting accommodates variations in thickness along a trees' length.
Branch connections tab
Largest gap: Type a value into the box to specify the largest "jump" distance to make a connection.
Max deviation angle: Type a value into the box to specify the maximum angle considered when connecting branches. The maximum value is 180°.
Min ratio of diameters: When two trees are considered for connection based on their respective location in (X, Y, Z) space, the diameters of the end point of each tree are compared. If the ratio of the diameters is greater than the specified value, the trees are connected and colored according to the most proximal tree.
Click in the slider to adjust.
Example:
The software compares the proximal end-point diameters of trees A and B.
- tree A diameter at end point (DA) > tree B diameter at end point (DB)
- The default ratio is 50%.
A and B will be connected if DB≥ (DA X 50%)