Register sections
Purpose

|
Use Register Sections to align images of individual serial sections or 3D reconstructions, created using the Assemble Serial Sections workflow, to a reference-brain atlas. The Register Sections tools are designed for images in which each image plane in the file was acquired independently, i.e., from sections on a slide. |
Use Register volume rather than Register Sections if you're working with 3D images obtained from cleared tissue, blockface images, or any tissue that was imaged whole.
See also Registration windows and tools reference.
Procedure
A. Getting started
-
If you are working with specimens from animals other than mice, download a brain atlas for the species from the NeuroInfo section of the MBF Bioscience download center.
When you install the brain atlas for your species, it will be available in the Atlas: dropdown menu.
-
Open an image of a single section or a 3D brain reconstruction image stack, for example, one created from images of serial sections using the Assemble Serial Sections workflow, using any of the following methods:
-
Drag and drop the file onto the NeuroInfo software window.
-
 Click Open an image file button in the Quick Access bar or type Ctrl+O.
Click Open an image file button in the Quick Access bar or type Ctrl+O. -
 Go to File > Open > Image Stack.
Go to File > Open > Image Stack.
-
-
 Click Register Sections in the NeuroInfo tools section of the Registration ribbon.
Click Register Sections in the NeuroInfo tools section of the Registration ribbon. -
The Register Section to Atlas window opens on the right; this is where you'll do most of the work.
-
The Registration ribbon displays functions relevant for registering sections to the atlas. Other tools are not displayed until you click
 Leave Registration.
Leave Registration.
-
-
If you are working with a 3D brain reconstruction image: Navigate to a section near the center of the brain that exhibits good contrast and distinguishable features, such as the hippocampus and fiber tracts, using the 3D Slice Scroll slider or the Orthoview tool.
-
 Using the Image Adjustment tool (available on the Workspace ribbon), we recommend making the following adjustments:
Using the Image Adjustment tool (available on the Workspace ribbon), we recommend making the following adjustments:-
Display only the color channel(s) with the anatomic information that you want to use for alignment. Deselect the other color channels so that they are not displayed.
-
Choose a light color, such as yellow, pale orange or pale grey to display the selected color channel(s).
-
Adjust the histogram so that structures in the image are clearly visible.
-
B. Select or create an atlas calibration [Current calibration section]
Context
Calibration provides NeuroInfo with enough information about how your section or 3D reconstruction relates to the reference-brain atlas to enable the effective use of automatic registration (alignment) tools. It establishes:
-
the brain atlas you are using as the reference
-
the anatomic orientation of the experimental image data
-
approximate relative scaling of the brain specimen(s) to the atlas
Do one of the following to set the calibration:
-
Select an appropriate calibration from the list. You can typically use the same calibration for registering brain specimens from the same species that were processed and imaged using equivalent protocols.
-
Create a new calibration:
-
the first time you use NeuroInfo
-
for specimens processed and/or imaged using a different protocol
-
to work with specimens from other species
-
-
Change an existing calibration: Select a calibration in the list and click Edit. Note that you will overwrite the existing calibration when you save the edited version.
-
 To create a new calibration, click Create New in the Calibrations section, under Current Calibrations.
To create a new calibration, click Create New in the Calibrations section, under Current Calibrations.-
A dialog box opens that prompts you to Enter Calibration Name. Type in a name that reflects the specimen and its processing method, then click OK.
-
You'll see the name for the new calibration at the top of the registration panel, Current Calibration: Your New Calibration.
 To edit an existing calibration, select a calibration and click Edit. [Note that if you edit a calibration, you will overwrite the existing calibration when you save the edited version.]
To edit an existing calibration, select a calibration and click Edit. [Note that if you edit a calibration, you will overwrite the existing calibration when you save the edited version.]Information to guide you through the atlas calibration process is displayed. Follow the steps outlined onscreen (and below)
-
-
Choose (or verify) the reference-brain atlas for the registration using the Atlas: drop-down menu
-
Allen2017Coronal_25um: Recommended for most registrations.
This atlas was created in the coronal orientation, with 25 µm sampling, by the Allen Institute for Brain Science in 2017.
-
GerfenNissl2017: You may want to choose this atlas if your specimen is labeled with a fluorescent dye that targets Nissl bodies (such as NeuroTrace dyes from ThermoFisher Scientific), and you are not satisfied with the precision of registration results using the Allen2017Coronal_25um atlas.
-
-
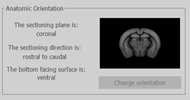 Check the Anatomic Orientation and click Change Orientation if the orientation of your 3D reconstruction is not accurately represented.
Check the Anatomic Orientation and click Change Orientation if the orientation of your 3D reconstruction is not accurately represented. The Orientation Selector dialog opens and prompts you to enter specimen-orientation information.
-
Expand the Registration Info section and review/update the Image Info:
-
Modality: select the microscopy mode used to process and image the experimental specimen.
-
Channel: Select the color channel with the cytoarchitectural information that will be used to align/register the experimental sections to the atlas.
-
-
Use the sliders in the Manual Registration Adjustment section to visually align the experimental and atlas images:
-
Click to expand the Manual Registration Adjustment and Visualization Options tools.
Working with a 3D brain reconstruction image: If you haven't already done so, use the 3D Slice Scroll slider or the Orthoview tool to navigate to a section in the experimental brain with good contrast and distinguishable features, such as the hippocampus and fiber tracts. Typically a section near the center of the brain is ideal.
Working with a single section: Click in the atlas brain slice view (bottom right) and
-
Move, scale and deform the atlas slice image so that it aligns with your experimental image slice:
-
Use the sliders in the Manual Registration Adjustment panel to Shift, adjust Rotation, and Scale the atlas slice to match the experimental slice image.
The sliders are labeled as follows:
-
D and V: dorsal and ventral
-
R and C: rostral and caudal
-
L and R: left and right
-
-
Note that one of the Shift sliders (which one depends on the specimen orientation) changes the plane (section) of the atlas displayed. Use it to display the section of the atlas brain that most closely matches the experimental section displayed.
-
It's helpful to turn the atlas overlay on and off as you align the images. Do this by toggling the checkbox and/or adjusting the slider to Show atlas over experiment slice in the Experimental Display panel.
-
-
-
 Press Run linear in the Registration section of the panel to refine the alignment between atlas and experimental section.
Press Run linear in the Registration section of the panel to refine the alignment between atlas and experimental section.If needed, you can repeat steps 5 and 6 to further refine the alignment.
-
Save the calibration when you are satisfied with this initial alignment/registration; click Save in the Calibrations section at the top of the registration tools panel.
-
Once saved, the calibration will be selected by default the next time that you open NeuroInfo; it can be used with other specimens from the same species that were prepared using the same tissue processing procedures.
-
C. Register (align) your experimental images to the atlas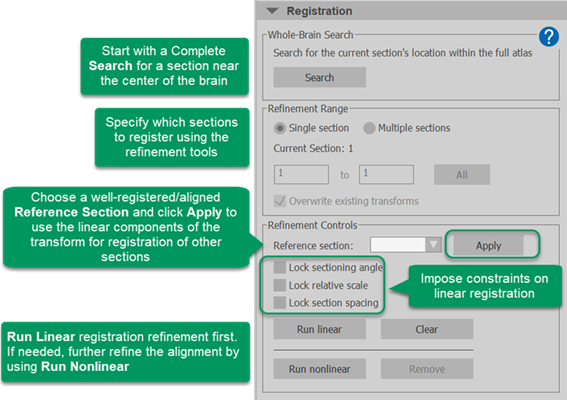
-
Use the Whole-Brain Search function to establish the atlas location, sectioning angle, and scale relative to the atlas for one to a few sections near the center of the brain as follows.
-
 With a section that exhibits good contrast and distinguishable features, Click Search in the Whole-Brain Search section of the panel to register (align) the current section to the atlas.
With a section that exhibits good contrast and distinguishable features, Click Search in the Whole-Brain Search section of the panel to register (align) the current section to the atlas.-
NeuroInfo searches the entire atlas to find the atlas location of the current section, then refines the alignment by determining the best optical sectioning angle.
-
When the search is complete, the atlas slice image should closely match the experimental section. You can use the controls in the Manual Registration Adjustment section to modify the alignment if desired.
-
-
You may want to register 2-4 sections across the center of the brain using the Whole-Brain Search.
-
-
Register additional sections or the entire 3D brain reconstruction to the atlas using the Refinement tools described below.
-
For each section or range of sections, we recommend that you Run Linear first, evaluate the precision of the alignment for each section individually, then repeat the Linear registration or click Run nonlinear and re-evaluate the alignment.
-
Repeat the registration of each section multiple times if desired. Running the registration algorithms multiple times will update and potentially further improve the registration result.
Refinement range:
-
Choose whether to register a Single section or Multiple sections.
-
The Current Section is displayed. Use the 3D slice scroll tool or the Orthoview tool to change the current section if needed.
-
Click All to select all sections or type in the section numbers you want to include (available when Multiple sections is selected above)
-
Overwrite existing transforms: Check to overwrite any existing section registrations when you run the refinement operation.
Refinement Controls:
-
Reference section: Select a well-aligned section that has already been registered from the dropdown menu; it will be referenced for both Linear and Nonlinear registration refinement of additional sections.
-
Click Apply to view the transform from the selected reference section on the current section.
These check boxes constrain Linear registration refinement only:
-
Lock sectioning angle: Check to hold the sectioning angle constant, using the angle identified for the reference section, while running a linear registration.
We recommend using this constraint in most cases; the sectioning angle identified near the center of the brain, where there is good contrast and multiple distinguishable features is typically more accurate than sectioning angle identified in parts of the brain with less contrast and more homogeneous anatomies.
-
Lock relative scale: Check to use the relative scale (compared to the atlas) identified for the reference section and keep the registration scale parameter constant while running a linear registration.
-
Lock section spacing: Check to maintain the section-spacing specified in the image file.

Run linear/Clear: Use linear registration, with the selected constraints to refine the alignment of the selected section(s) / Clear the linear registration results

Run nonlinear/Remove: Use nonlinear registration to refine the alignment of the selected section(s) / Clear the nonlinear registration results.
-
-
Save your work.
-
 Save the data file by clicking Save in the quick access menu, typing Ctrl+S, or by going to File > Save > Data file.
Save the data file by clicking Save in the quick access menu, typing Ctrl+S, or by going to File > Save > Data file. -
 >
>  Save your transform set by clicking Manage transforms on the Registration ribbon, then Save transform set from the dropdown menu. The transform set includes atlas-alignment information for each section that was registered.
Save your transform set by clicking Manage transforms on the Registration ribbon, then Save transform set from the dropdown menu. The transform set includes atlas-alignment information for each section that was registered.
-
C. Identifying anatomical structures
Once you have registered (aligned) one or more sections to a reference atlas, you can identify atlas structures in your experimental image(s) as follows:
-
See the name of the anatomical structure at the cursor location: Hover your mouse over the Experimental or Atlas Slice images (top and bottom images on the right) to display the name of the brain structure in the lower left corner of the image panel.
-
Display the outline of the anatomical feature:
-
Ctrl + click the location with your mouse
-
 Click Select Anatomy in the Registration ribbon and click the location with your mouse.
Click Select Anatomy in the Registration ribbon and click the location with your mouse.  Click Deselect Anatomy to turn off display of anatomical structures.
Click Deselect Anatomy to turn off display of anatomical structures. -
 Click the check box for the structure name in the list displayed on the Atlas Ontology tab to display its outline. Uncheck the box to stop its display.
Click the check box for the structure name in the list displayed on the Atlas Ontology tab to display its outline. Uncheck the box to stop its display.
-
Learn more about identifying anatomical structures and using the Atlas Ontology tools.
Learn about Mapping experiments to/from the atlas