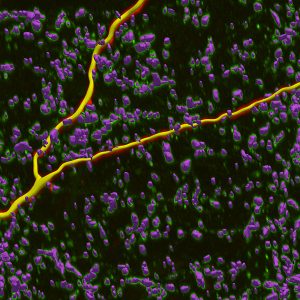
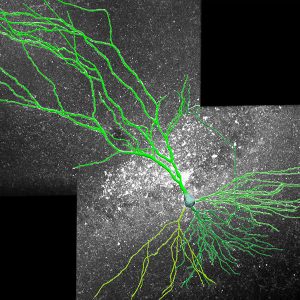
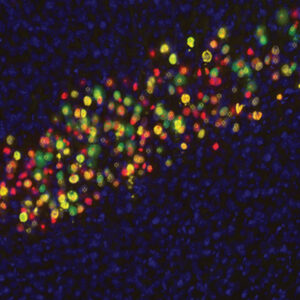
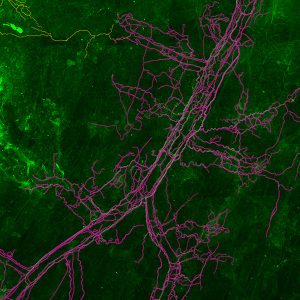
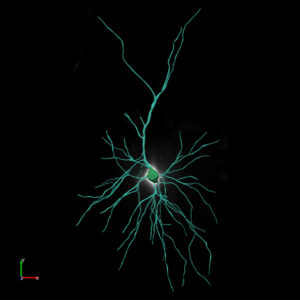
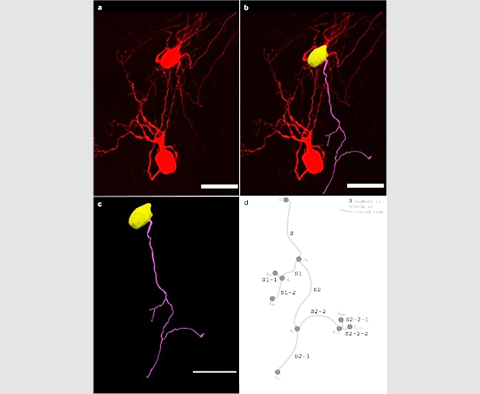
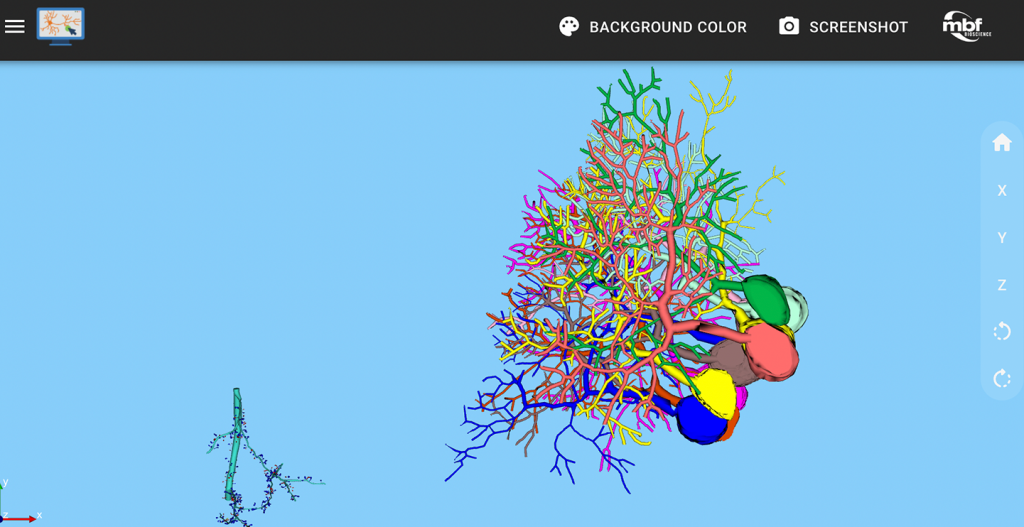
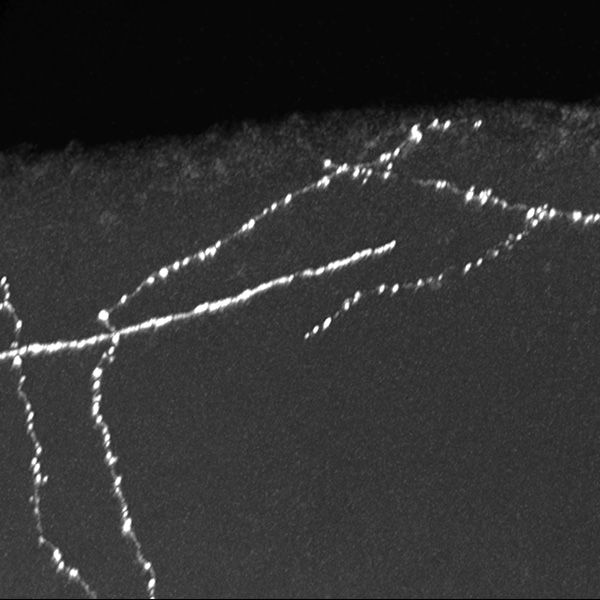
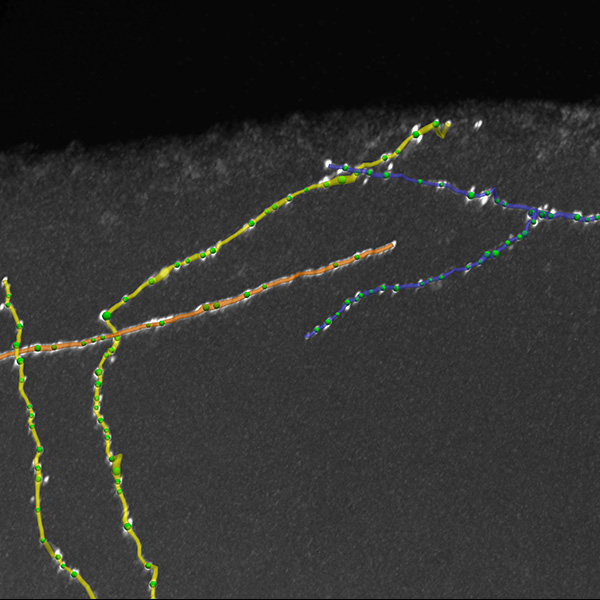
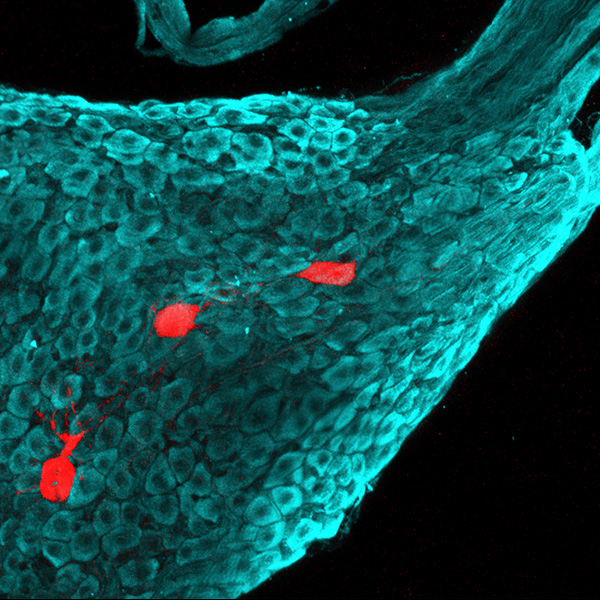
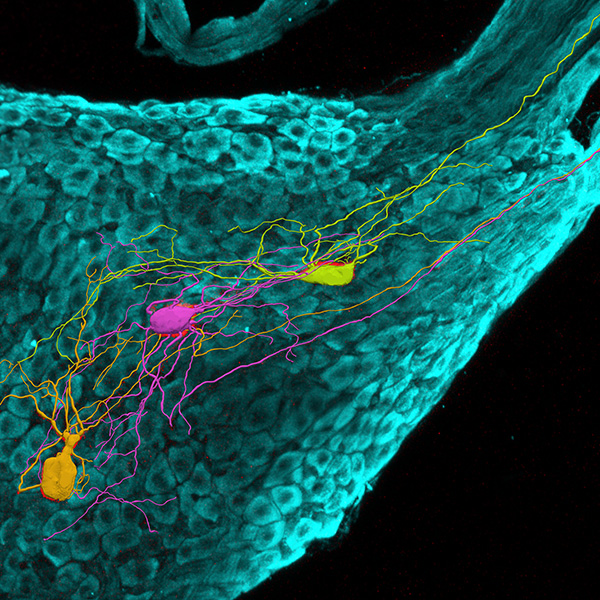
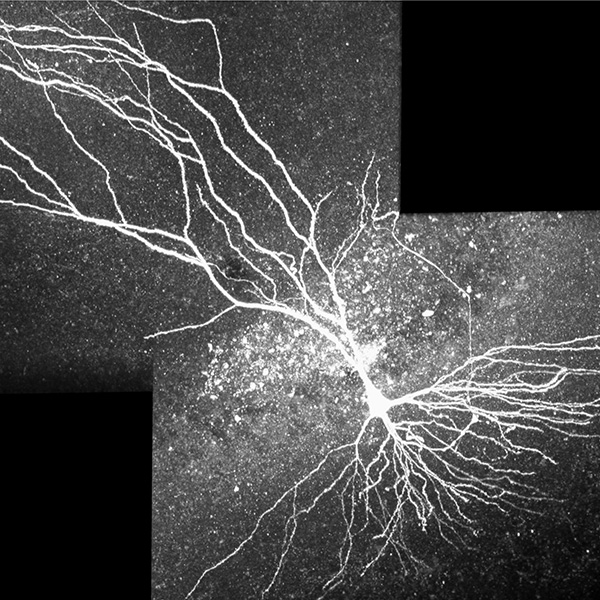
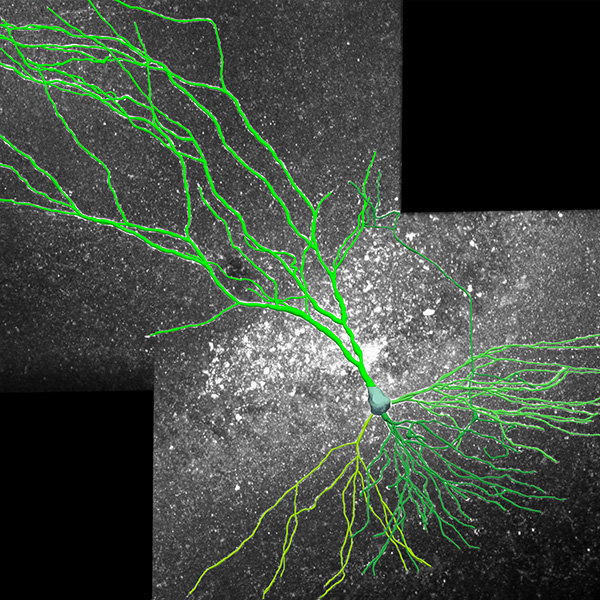
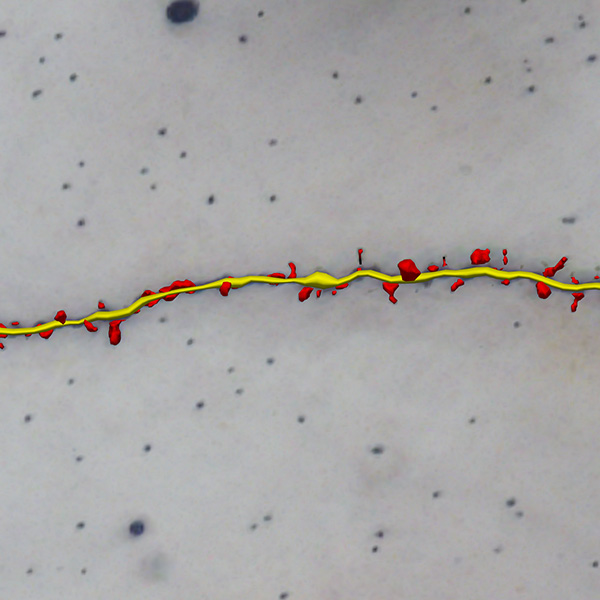
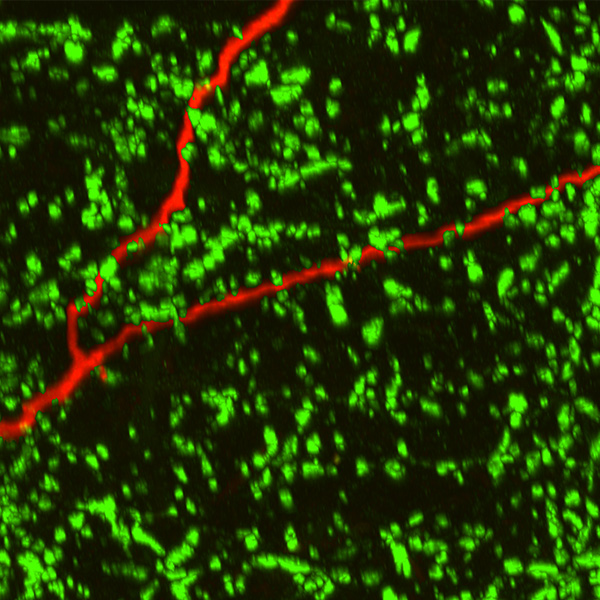
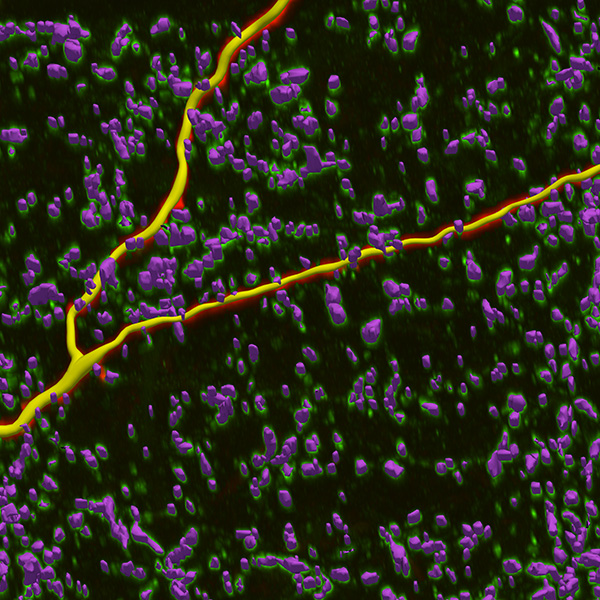
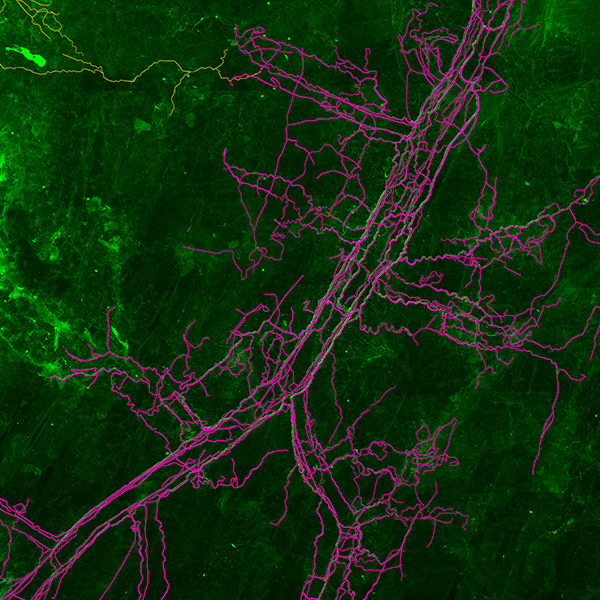
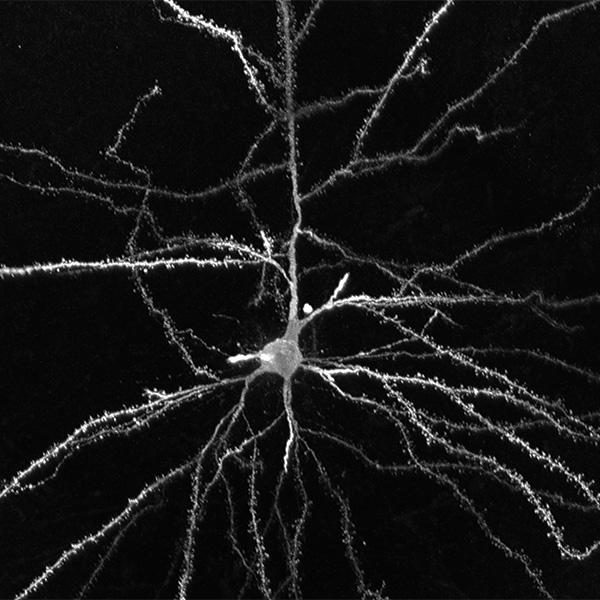
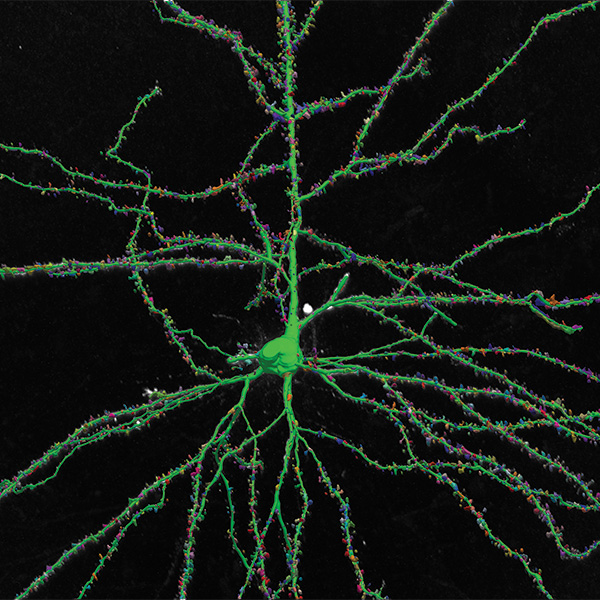
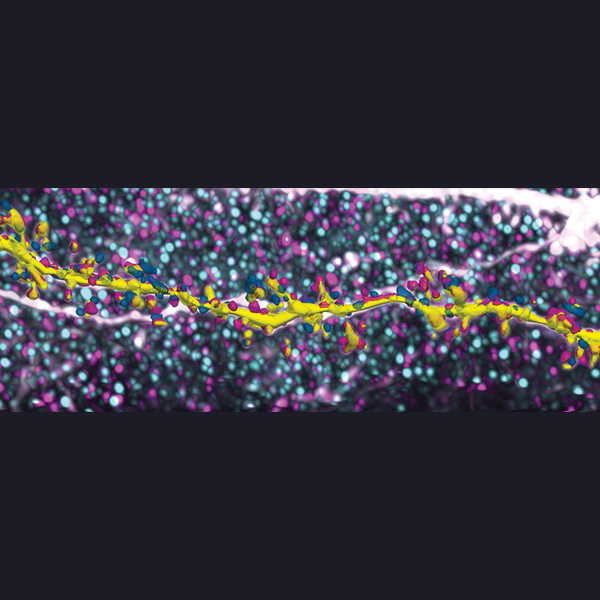
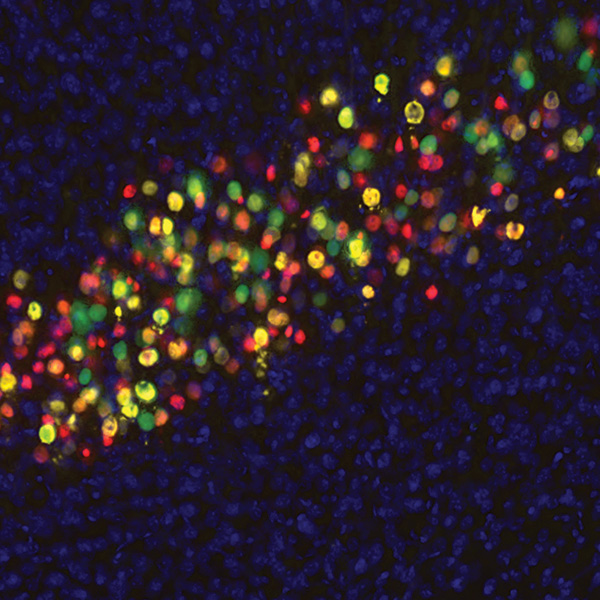
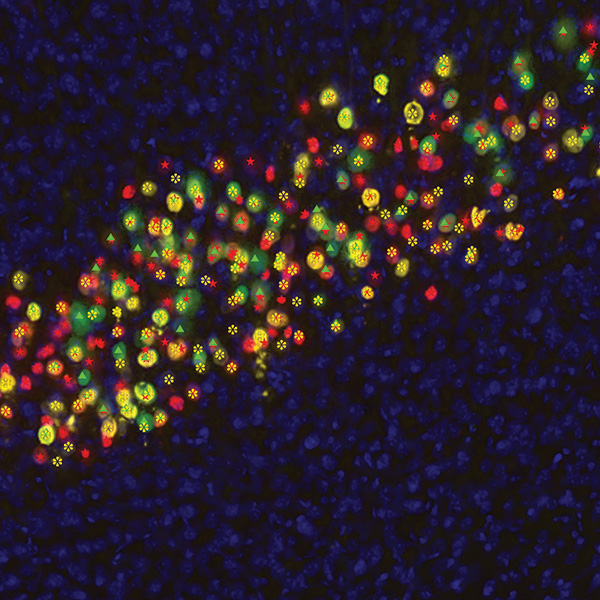
Neurolucida 360: The leading image analysis software for automatic 3D neuron tracing and reconstruction and quantitative morphology
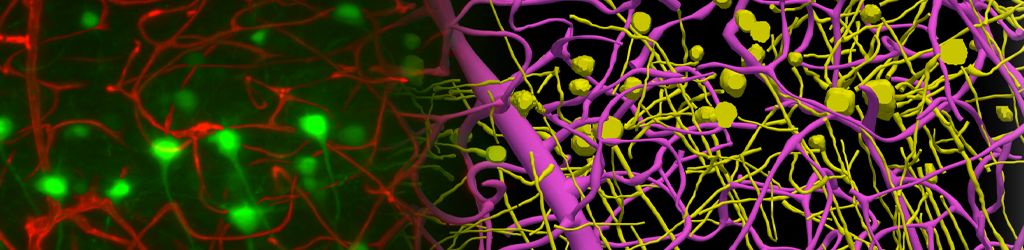
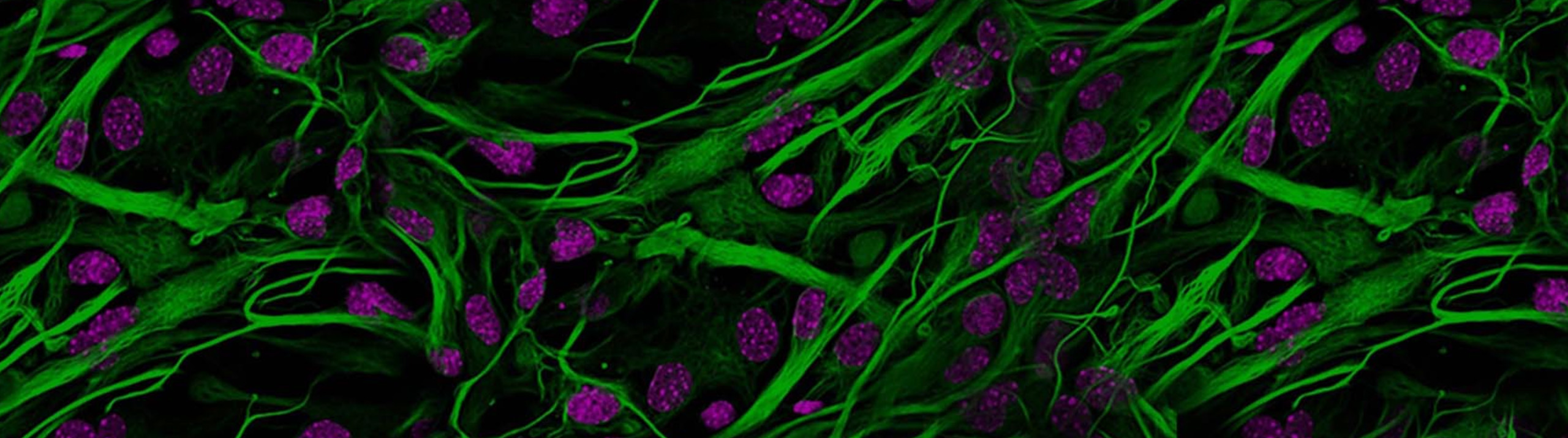
Neurolucida 360 is the most trusted software for comprehensive neuron tracing and reconstruction. Built by neuroscientists for neuroscientists, Neurolucida 360 is equipped with powerful automated algorithms for detection of neuronal structures present in 2D and 3D microscopy image data.
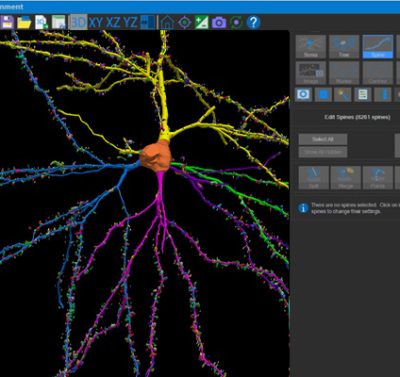
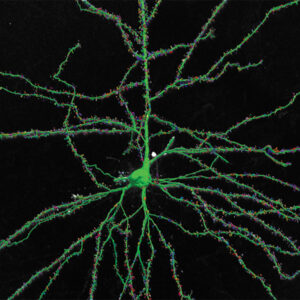
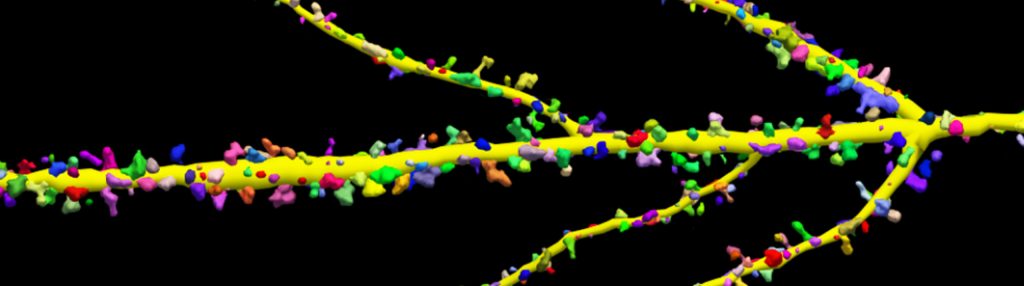
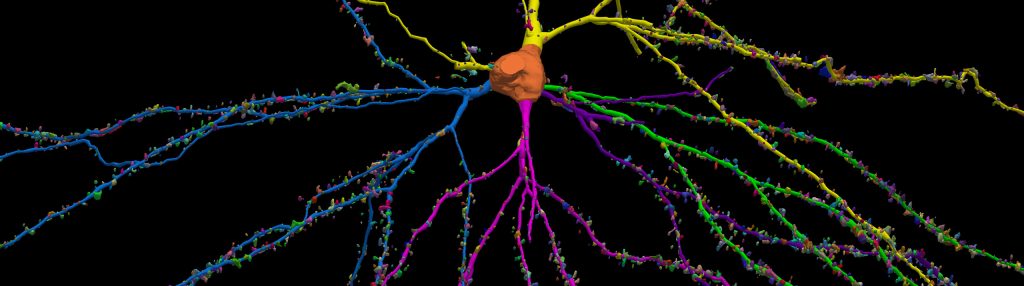
With built-in workflows, an intuitive user-interface, and interactive 3D environment, Neurolucida 360 makes it easy to reconstruct somas, axons, dendrites, varicosities, spines, and synapses.
Paired with the companion software, Neurolucida Explorer, you can quickly and effectively analyze the morphology of subcellular structures, whole cells, or complex connective networks – from any species.
Neurolucida 360 supports many types of histological preparation techniques and microscopy modalities, such as:
- Immunofluorescence
- Golgi-Cox
- AAVs
- Retrograde or anterograde filled cells
- Immunohistochemistry
- Brightfield
- Widefield fluorescence
- Confocal
- Light sheet
- 2-Photon, Multi-Photon
- Whole brain or organ
- Cleared tissue
- Sectioned tissue
- Cell cultures
- Expanded tissue
Neurolucida 360 has been developed with support from the National Institute of Mental Health (NIMH)
Ultra is here! A 3D automatic vessel tracing module for thorough exploration of neuro-vascular environments.
Recommended Hardware Requirements
Neurolucida 360 is a powerful software that can work with a wide range of image data of vastly different levels of size and complexity. Because of its flexibility, our recommendations for system configurations vary to balance the affordability of lower computing power for smaller data sets to more expensive high-end workstations for large, multi-channel data.
Please contact us to consult with our technical specialists for a more precise recommendation specific to your data needs.
| 64-bit Windows 11 operating system |
| CPU with 8 cores (16 threads) or more. More cores improve performance when using large data sets (>1 GB). |
| Solid state hard drive(s). Preferably, non-volatile memory express (NVMe) drives. |
| 64 GB of system memory or more. More memory is better for large data sets, especially for image handling and reconstruction in the 3D environment. |
| Graphics card with 8 GB memory or more. Most graphics cards from NVIDIA and AMD have been tested with MBF Bioscience software. |
| Minimum Hardware Requirements |
| 64-bit Windows 10 operating system |
| 8 core processor (16 threads) |
| 32 GB memory |
| Graphics card with 6 GB memory or more: This is the minimum needed for the 3D environment, but is adequate for working only in 2D. |
| Computer-Hardware Upgrade Priorities |
| To upgrade your system for better performance with MBF Bioscience software, we suggest that you prioritize computer hardware upgrades as follows: |
-
-
- Increase system memory to at least 128 GB. More memory improves performance when using large data sets
|
-
-
- Replace hard disk drives (HDDs) with solid-state drives (SSDs), preferably non-volatile memory express (NVMe) drives
|
-
-
- If you're working with 3D images and graphics, upgrade graphics card (GPU) to one with at least 8-12 GB memory
|
From Proteins to Dendritic Spines: Neurolucida 360 Plays a Crucial Role in Advancing Neuroscience
>> Learn More
Blue Brain scientists develop pyramidal cell classification system using neurons reconstructed with Neurolucida 360
>> Learn More
Curcumin Lowers Neuroinflammation in Mouse Model
>> Learn More
Researchers Identify Potential Treatment for Patients at Risk for Alzheimer’s Disease
>> Learn More
MBF Bioscience research team contributes novel dendritic spine analysis in study published in Science
>> Learn More
NeuroMorpho.Org Releases Nearly 10,000 New Neuron Reconstructions and Neurolucida leads the way
>> Learn More
Visit our blog to read more case studies
Download Neurolucida 360 product sheet here.
Neurolucida® 360 Version 2024.2.2
Released December 2024
Licensing Update
Update to offline software licensing. Neurolucida 360 software requires reactivation on systems that do not have access to the internet. Instructions for offline software activation are included in the Neurolucida 360 user guide.
New Features and Enhancements
-
- Improvements to the vessel tracing options in the 2D window
- Automove will now reposition focus when Continuous tracing is being used
- Color channel selection in puncta setup will begin at number 1
- Improvements to synchronization of contours delineated in 2D and 3D
- Image file paths are displayed in the Image Organizer
- During a movie, you can now change image-display settings for image slice and partial projections
- New ability to display the 3D scale bar while recording a Movie
- Other improvements and updates to the Movie mode interface have been implemented
See a full list of new features here
An AI-Extended Solution for Performing Integrated 3D Neuro Microstructure and Nanostructure Analysis of Expansion Microscopy Data